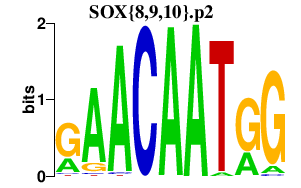
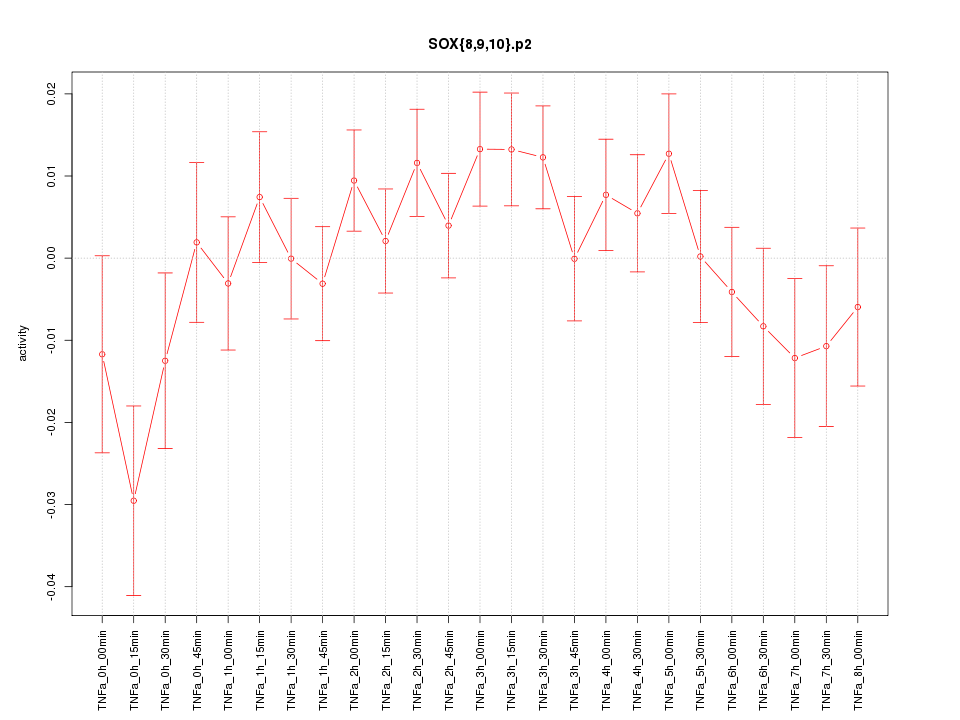
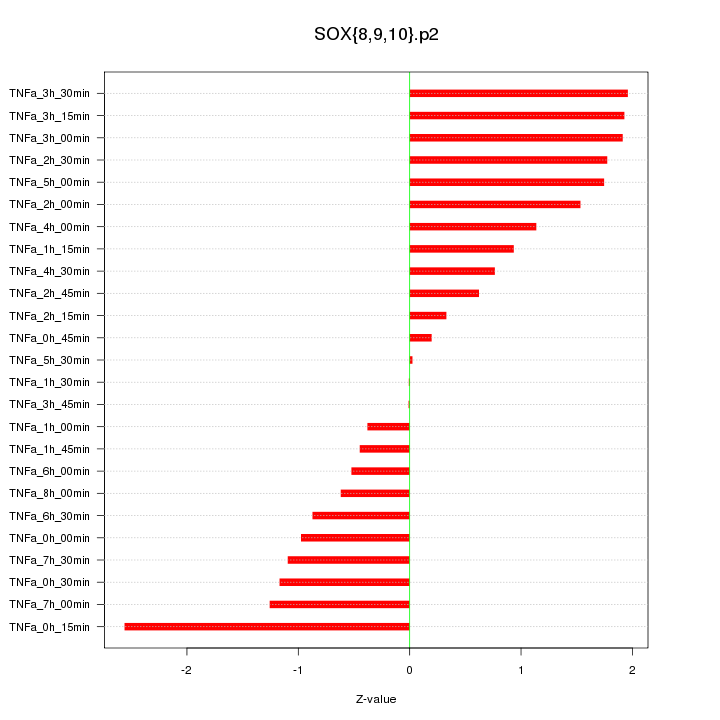
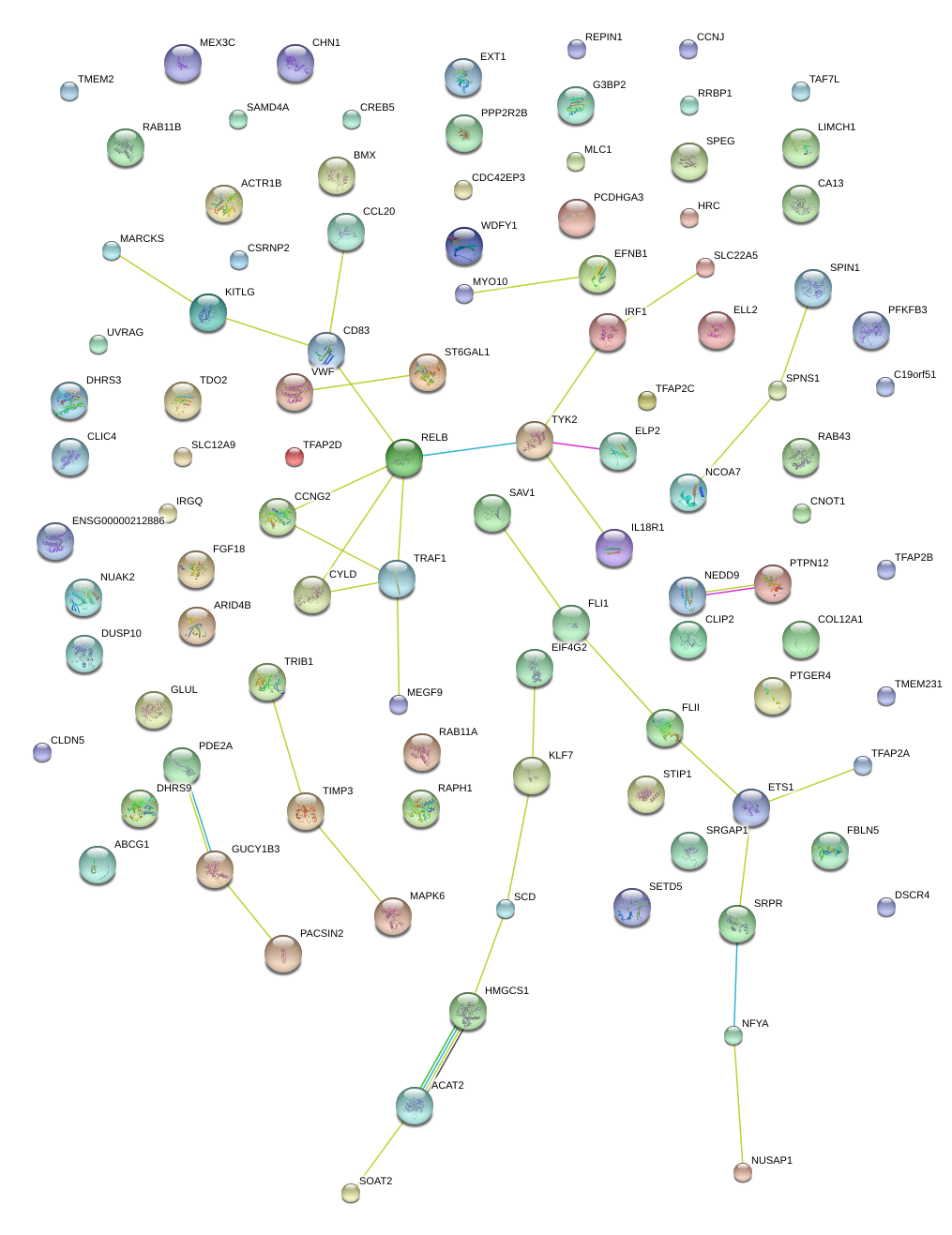
|
chr6_-_11232900
|
3.907
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr21_+_26934440
|
3.207
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr5_-_131826426
|
3.052
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr19_+_45504686
|
2.796
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr6_-_11232883
|
2.570
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr10_+_6244839
|
2.472
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr1_-_205290756
|
2.463
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr6_-_11232901
|
2.384
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr2_+_228678556
|
2.211
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr19_-_49658645
|
2.171
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr15_+_52311438
|
2.160
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr3_+_186648515
|
2.132
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr6_-_10415438
|
2.112
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr2_-_208031570
|
2.052
|
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr21_+_43710190
|
2.018
|
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr5_-_16936290
|
2.012
|
|
MYO10
|
myosin X
|
|
chr14_-_51135021
|
1.932
|
NM_021818
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr16_-_75590107
|
1.915
|
NM_001077416
NM_001077418
NM_001077419
|
TMEM231
|
transmembrane protein 231
|
|
chr15_+_52311432
|
1.908
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr22_-_19512859
|
1.886
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr6_+_14117486
|
1.832
|
NM_001251901
|
CD83
|
CD83 molecule
|
|
chr15_+_52311408
|
1.796
|
NM_002748
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr4_+_156680125
|
1.745
|
NM_000857
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr7_+_77167351
|
1.725
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr2_+_102979096
|
1.651
|
NM_003855
|
IL18R1
|
interleukin 18 receptor 1
|
|
chr4_+_78079302
|
1.632
|
|
CCNG2
|
cyclin G2
|
|
chr7_+_73703704
|
1.574
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr11_+_128563955
|
1.537
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr3_+_9439533
|
1.508
|
|
SETD5
|
SET domain containing 5
|
|
chr9_-_123689172
|
1.485
|
NM_005658
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr20_-_17662704
|
1.484
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr9_-_123476578
|
1.474
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr1_-_182360181
|
1.457
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr7_+_150065968
|
1.447
|
|
REPIN1
|
replication initiator 1
|
|
chr11_-_126138789
|
1.429
|
|
SRPR
|
signal recognition particle receptor (docking protein)
|
|
chr4_-_76598558
|
1.410
|
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr11_-_10830034
|
1.406
|
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr6_-_75915566
|
1.366
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr14_+_55168831
|
1.349
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr1_-_235491448
|
1.323
|
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr11_+_63953630
|
1.320
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr18_-_48723689
|
1.305
|
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr1_-_12677347
|
1.287
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr9_+_91003987
|
1.282
|
|
SPIN1
|
spindlin 1
|
|
chr1_-_221915426
|
1.269
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr14_-_51134862
|
1.254
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr14_-_51135041
|
1.252
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr16_+_50776012
|
1.243
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr1_-_182357757
|
1.227
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr16_+_50775960
|
1.215
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr19_-_10491172
|
1.213
|
|
TYK2
|
tyrosine kinase 2
|
|
chr22_+_33197678
|
1.191
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr8_-_119123804
|
1.170
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr19_+_8455243
|
1.164
|
|
RAB11B
|
RAB11B, member RAS oncogene family
|
|
chr19_+_8455265
|
1.162
|
|
RAB11B
|
RAB11B, member RAS oncogene family
|
|
chr20_-_17662828
|
1.158
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr5_+_140739589
|
1.155
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr12_-_88974094
|
1.144
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr19_-_10491212
|
1.133
|
NM_003331
|
TYK2
|
tyrosine kinase 2
|
|
chrX_+_15518899
|
1.097
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr12_-_6233684
|
1.081
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr6_+_126240369
|
1.075
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr16_-_58663727
|
1.058
|
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr8_+_126442557
|
1.058
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chrX_+_68048792
|
1.041
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr2_-_37899227
|
1.021
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr2_-_37898768
|
1.018
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr22_+_33197744
|
1.018
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr9_-_74383301
|
1.004
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr11_+_75526236
|
0.995
|
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr5_+_140797243
|
0.995
|
NM_018927
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr11_+_128563993
|
0.991
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr6_+_160183510
|
0.980
|
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr11_+_63953666
|
0.970
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr7_+_28452143
|
0.966
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr10_+_102106771
|
0.961
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chrY_-_21239250
|
0.957
|
|
TTTY14
|
testis-specific transcript, Y-linked 14 (non-protein coding)
|
|
chr3_-_9439122
|
0.957
|
|
LOC440944
|
uncharacterized LOC440944
|
|
chr7_+_100450327
|
0.957
|
|
SLC12A9
|
solute carrier family 12 (potassium/chloride transporters), member 9
|
|
chr6_+_41040713
|
0.952
|
|
NFYA
|
nuclear transcription factor Y, alpha
|
|
chr11_-_72380107
|
0.944
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr1_+_25071906
|
0.944
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr12_+_64238511
|
0.941
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr22_-_50523780
|
0.936
|
NM_139202
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr11_+_128564130
|
0.932
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr22_-_50523737
|
0.931
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr15_+_41625152
|
0.931
|
|
NUSAP1
|
nucleolar and spindle associated protein 1
|
|
chr12_-_51477138
|
0.930
|
|
CSRNP2
|
cysteine-serine-rich nuclear protein 2
|
|
chr5_-_43313492
|
0.918
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr6_+_114178516
|
0.913
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr22_-_50523853
|
0.913
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr22_-_43343046
|
0.912
|
NM_007229
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr19_-_55677919
|
0.910
|
NM_178837
|
C19orf51
|
chromosome 19 open reading frame 51
|
|
chr2_-_54198055
|
0.909
|
|
|
|
|
chr5_-_95297424
|
0.907
|
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr8_+_86157693
|
0.905
|
NM_198584
|
CA13
|
carbonic anhydrase XIII
|
|
chr2_-_175869910
|
0.905
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr2_-_204399906
|
0.898
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr14_-_92413738
|
0.892
|
|
FBLN5
|
fibulin 5
|
|
chr5_+_140797270
|
0.882
|
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr11_+_128563973
|
0.879
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr11_+_75526211
|
0.878
|
NM_003369
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr11_-_126138698
|
0.872
|
|
SRPR
|
signal recognition particle receptor (docking protein)
|
|
chr19_-_44100127
|
0.865
|
NM_001007561
|
IRGQ
|
immunity-related GTPase family, Q
|
|
chr5_+_40680042
|
0.864
|
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr11_-_128457452
|
0.863
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr5_+_140729723
|
0.861
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr5_-_146435589
|
0.859
|
NM_181674
NM_181676
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr2_-_224810020
|
0.850
|
|
WDFY1
|
WD repeat and FYVE domain containing 1
|
|
chr2_+_169923531
|
0.847
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr5_+_170846659
|
0.840
|
NM_003862
|
FGF18
|
fibroblast growth factor 18
|
|
chr4_-_76598563
|
0.833
|
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr2_+_220309373
|
0.832
|
|
SPEG
|
SPEG complex locus
|
|
chr1_+_25071967
|
0.829
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr11_+_128563772
|
0.826
|
NM_001167681
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr10_+_97803064
|
0.818
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr4_+_41614911
|
0.806
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr14_+_63671101
|
0.803
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr9_+_116263706
|
0.800
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr2_+_170590454
|
0.797
|
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr1_-_55352893
|
0.795
|
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chrX_+_41193409
|
0.794
|
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked
|
|
chr9_-_139658959
|
0.787
|
NM_203347
|
LCN15
|
lipocalin 15
|
|
chr11_-_57417247
|
0.783
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr12_+_110011616
|
0.781
|
|
MVK
|
mevalonate kinase
|
|
chr8_-_38325524
|
0.781
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr11_+_63953586
|
0.779
|
NM_006819
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr11_+_67007488
|
0.776
|
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr3_+_196466806
|
0.776
|
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chrX_-_45060081
|
0.774
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr22_-_50524357
|
0.772
|
NM_015166
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr1_-_95392449
|
0.771
|
|
CNN3
|
calponin 3, acidic
|
|
chr16_-_84538219
|
0.761
|
NM_020947
|
KIAA1609
|
KIAA1609
|
|
chr17_-_49124128
|
0.754
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr8_-_6420454
|
0.753
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr4_+_41614933
|
0.750
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr19_+_57106622
|
0.747
|
NM_021216
|
ZNF71
|
zinc finger protein 71
|
|
chr5_+_74633043
|
0.745
|
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase
|
|
chr11_+_129991582
|
0.741
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr12_+_108909049
|
0.736
|
NM_007076
|
FICD
|
FIC domain containing
|
|
chr1_-_221915458
|
0.735
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr14_-_55369163
|
0.733
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr1_-_55352920
|
0.727
|
NM_014762
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr1_-_235491494
|
0.726
|
NM_016374
NM_031371
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr11_-_126138824
|
0.723
|
NM_001177842
NM_003139
|
SRPR
|
signal recognition particle receptor (docking protein)
|
|
chr6_-_30655078
|
0.722
|
NM_001134870
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr8_-_101965096
|
0.715
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr16_+_67063214
|
0.714
|
|
CBFB
|
core-binding factor, beta subunit
|
|
chr22_-_43411129
|
0.713
|
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr1_-_55352818
|
0.713
|
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr15_+_85923739
|
0.709
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr1_-_221915393
|
0.708
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr2_-_224810015
|
0.706
|
|
WDFY1
|
WD repeat and FYVE domain containing 1
|
|
chr3_-_160283071
|
0.705
|
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr3_+_44690216
|
0.700
|
NM_003420
|
ZNF35
|
zinc finger protein 35
|
|
chr16_-_84651519
|
0.699
|
|
COTL1
|
coactosin-like 1 (Dictyostelium)
|
|
chr10_+_18948276
|
0.694
|
NM_178815
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr3_-_160283166
|
0.686
|
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr6_+_26383323
|
0.685
|
NM_001197238
NM_001197239
NM_001197240
NM_006995
NM_181531
NM_001197237
|
BTN2A2
|
butyrophilin, subfamily 2, member A2
|
|
chr6_+_83072922
|
0.679
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chr2_-_224810041
|
0.678
|
NM_020830
|
WDFY1
|
WD repeat and FYVE domain containing 1
|
|
chr1_-_55352881
|
0.673
|
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr7_-_11871735
|
0.671
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr6_-_107435635
|
0.668
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr6_+_160182988
|
0.667
|
NM_005891
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr2_-_158182154
|
0.667
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr2_+_159313407
|
0.666
|
|
PKP4
|
plakophilin 4
|
|
chr7_-_75368276
|
0.666
|
NM_001243198
NM_005338
|
HIP1
|
huntingtin interacting protein 1
|
|
chr9_+_92219926
|
0.664
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr6_+_160183062
|
0.663
|
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr5_+_113698015
|
0.661
|
NM_021614
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr18_+_55888799
|
0.659
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr3_-_71353910
|
0.653
|
NM_001244812
|
FOXP1
|
forkhead box P1
|
|
chr6_-_32163890
|
0.649
|
|
NOTCH4
|
notch 4
|
|
chr7_-_75368216
|
0.648
|
|
HIP1
|
huntingtin interacting protein 1
|
|
chr19_-_47734215
|
0.647
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr6_-_30654978
|
0.645
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr6_+_30688085
|
0.643
|
|
TUBB
|
tubulin, beta class I
|
|
chr19_+_799281
|
0.642
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr8_-_82395401
|
0.638
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr14_+_59655351
|
0.638
|
NM_014992
|
DAAM1
|
dishevelled associated activator of morphogenesis 1
|
|
chr11_-_69519105
|
0.634
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr1_-_85930610
|
0.628
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr5_-_43313581
|
0.626
|
NM_001098272
NM_002130
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr1_-_153643433
|
0.623
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr18_+_55888838
|
0.621
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr14_+_103589711
|
0.619
|
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr2_-_43453724
|
0.616
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr4_-_101439071
|
0.616
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr7_+_5632453
|
0.615
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr14_+_103800534
|
0.615
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr1_-_153643456
|
0.614
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr14_-_91884164
|
0.613
|
NM_001080414
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr4_+_41540320
|
0.607
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr16_-_22385596
|
0.607
|
NM_001802
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa
|
|
chr2_+_189839132
|
0.606
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr14_-_73453651
|
0.605
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr19_+_42580333
|
0.604
|
|
ZNF574
|
zinc finger protein 574
|
|
chr1_-_153643428
|
0.603
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr19_+_42580275
|
0.602
|
NM_022752
|
ZNF574
|
zinc finger protein 574
|